Genes, peoples, and languages
L. LUCA CAVALLI-SFORZA
Department of Genetics, School of Medicine, Stanford University, Stanford, CA 94305-5120Abstract
The genetic history of a group of populations is usually analyzed by reconstructing a tree of their origins. Reliability of the reconstruction depends on the validity of the hypothesis that genetic differentiation of the populations is mostly due to population fissions followed by independent evolution. If necessary, adjustment for major population admixtures can be made. Dating the fissions requires comparisons with paleoanthropological and paleontological dates, which are few and uncertain. A method of absolute genetic dating recently introduced uses mutation rates as molecular clocks; it was applied to human evolution using microsatellites, which have a sufficiently high mutation rate. Results are comparable with those of other methods and agree with a recent expansion of modern humans from Africa. An alternative method of analysis, useful when there is adequate geographic coverage of regions, is the geographic study of frequencies of alleles or haplotypes. As in the case of trees, it is necessary to summarize data from many loci for conclusions to be acceptable. Results must be independent from the loci used. Multivariate analyses like principal components or multidimensional scaling reveal a number of hidden patterns and evaluate their relative importance. Most patterns found in the analysis of human living populations are likely to be consequences of demographic expansions, determined by technological developments affecting food availability, transportation, or military power. During such expansions, both genes and languages are spread to potentially vast areas. In principle, this tends to create a correlation between the respective evolutionary trees. The correlation is usually positive and often remarkably high. It can be decreased or hidden by phenomena of language replacement and also of gene replacement, usually partial, due to gene flow.
Which contains the
One reasonable hypothesis is that the genetic distance between Asia and Africa is shorter than that between Africa and the other continents in Table 1 because both Africans and Asians contributed to the settlement of Europe, which began about 40,000 years ago. It seems very reasonable to assume that both continents nearest to Europe contributed to its settlement, even if perhaps at different times and maybe repeatedly. It is reassuring that the analysis of other markers also consistently gives the same results in this case. Moreover, a specific evolutionary model tested, i.e., that Europe is formed by contributions from Asia and Africa, fits the distance matrix perfectly (6). In this simplified model, the migrations postulated to have populated Europe are estimated to have occurred at an early date (30,000 years ago), but it is impossible to distinguish, on the basis of these data, this model from that of several migrations at different times. The overall contributions from Asia and Africa were estimated to be around two-thirds and one-third, respectively.
Which doesn’t seem to fit the mt/Y DNA patterns, although to be fair L mt types don’t seem to thrive in a cold climate. Since he gives a 146,000 ya date for the first migration out of Africa, this second wave of expansion could have been a very long time ago. Possibly a double OOA might explain the total failure of Y chr dates to tally with the mt DNA expansion dates.
The first estimate gave a separation time of the first migrants out of Africa of 146,000 years ago, very close to the date obtained with the mtDNA full sequence. This was based on results with 30 microsatellites (5). More recent results (L. Jin, unpublished work) with 100 microsatellites gave an earlier date.
Also more humourously, but unlikely..
The Ethiopians genotype is more than 50% African. It is difficult to say if they originated in Arabia and are therefore Caucasoids who, like Lapps, had substantial gene flow after they migrated to East Africa, or if they originated in Africa and had substantial gene flow from Arabia, but not enough to pass the 50% mark.
I think the ‘ mixed expansion south from Egypt with some later Neolithic Arabian farmer’ is a more likely scenario.
I’ll admit to not reading the whole thing before posting it. I have a rotten headache and the kids are playing up. I’ll read it tomorrow.
And having had another look..
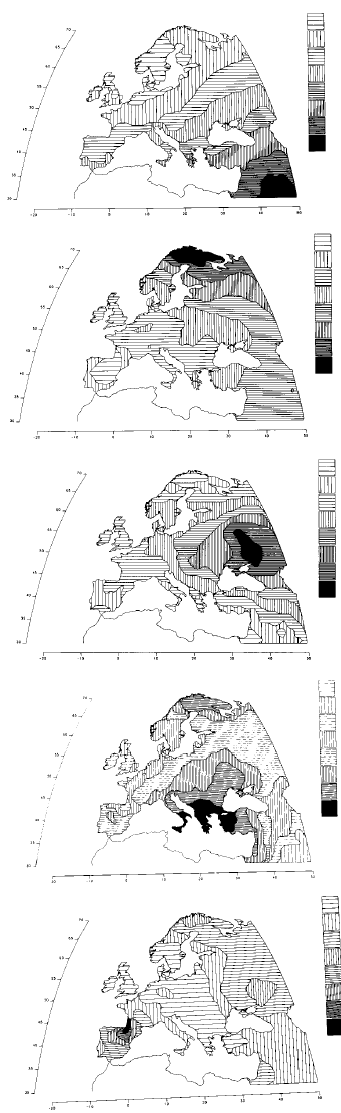
There’s this interesting map showing patterns of variation in Europe.
FIG. 2. Hidden patterns in the geography of Europe shown by the first five principal components, explaining respectively 28%, 22%, 11%, 7%, and 5% of the total genetic variation for 95 classical polymorphisms (1, 13, 14).
The first component is almost superimposable to the archaeological dates of the spread of farming from the Middle East between 10,000 and 6,000 years ago.
The second principal component parallels a probable spread of Uralic people and/or languages to the northeast of Europe.
The third is very similar to the spread of pastoral nomads (and their successors) who domesticated the horse in the steppe towards the end of the farming expansion, and are believed by some archaeologists and linguists to have spread most Indo-European languages to Europe.
The fourth is strongly reminiscent of Greek colonization in the first millennium B.C.
The fifth corresponds to the progressive retreat of the boundary of the Basque language. Basques have retained, in addition to their language, believed to be descended from an original language spoken in Europe, some of their original genetic characteristics. (From ref. 1, with permission of Princeton University Press, modified.)

I’m sure Luis will recognise these maps. I’ve used them in my series of essays at remotecentral.
The first principal component is my map in “The Human Star”, the second is in “Culture”, the third in “Indo-Europeans”, the fourth and fifth in “The Last Point”.
I’d be intersted in hearing his comments regarding these original versions.
Some intersting points.
“The first component is almost superimposable to the archaeological dates of the spread of farming from the Middle East”. But is just as likely to be the long term result of that along with expansion from around the North and Baltic Seas.
“The second principal component parallels a probable spread of Uralic people and/or languages to the northeast of Europe”. So the Uralic people spread in significant numbers southward between the Carpathian and Sudetan Mountains?
“The fifth corresponds to the progressive retreat of the boundary of the Basque language”. Perhaps, but let’s look at why it’s in retreat. Again an expansion from around the North and Baltic Seas?
Cavalli-Sforza has some more similar maps for other regions of the world in his “History and Geography of Human Genes”. I haven’t been able to find any of them on the net. Perhaps you might be able to Mathilda?
Not that I could find.
Thanks for trying.
The links just for the hell of it:
First component, my map 2 in the section ‘A Cline’:
http://remotecentral.blogspot.com/search/label/Human%20Evolution%20On%20Trial%20-%20Human%20Star
Second, my map 18 in the section ‘Europe’:
http://remotecentral.blogspot.com/search/label/Human%20Evolution%20on%20Trial%20-%20Culture
Third, map 7 in ‘Indo-European Languages’:
http://remotecentral.blogspot.com/search/label/Human%20Evolution%20On%20Trial%20-%20Indo-Europeans
The fourth, fifth and sixth (not shown in this post but available in Cavalli-Sforza’s book) make up maps 22-25 (I break the fourth component into two) scattered through:
http://remotecentral.blogspot.com/search/label/Human%20Evolution%20on%20Trial%20-%20Human%20Star%20-%20The%20Last%20Point
Terry: Autosomal PCs appear to be very susceptible to sampling and stuff like that. It’s quite interesting that CV in his book (not really a paper but a divulgation book, one of my first readings in genetics back there in the 90s, btw) finds a “Basque” component but, when you compare with other more recent studies, you realize that there are components (clusters) he did not detect (for instance a Central-North European one or an Iberian, non-Basque, one as well).
Also his attribution of some of the components appears misleading:
·PC1 and PC4: PC1 is attributed happily to Neolithic but in fact it hardly corresponds with it. PC4 instead does pretty well. PC1 might be Neolithic too but it appears to dominate areas where it’s not so strong actually, like Spain. PC4 in any case ahrdly relates to “Greek colonization” as Greeks never colonized the inland Balcans at all. That pattern appears much more closely related with Neolithic expansion also centered in Greece (Thessaly to be specific).
·PC2 and PC3: PC2 can ahrdly be considered Uralic once you look at the shadowed area in West Asia, where Uralic presence is unheard of. It rather seems to be some other kind of component, carried by Uralic peoples, sure, but also by other ethnicities, like Indo-Europeans maybe. The fact that it is much more important in terms of weight than PC3 also suggests that.
Anyhow PC1 and PC2 appear again in other studies (with some modifications) but when you look at deeper levels, they tend to become very thin in the West of Europe. They only appear that way because the Western diversity is greater and therefore the Western components appear more localized and weight less in the overall picture.
Check Bauchet et al, 2007 for a contrast. There you can see (K-means structure) how specific Central-North European and Iberian components appear and actually make up the biggest share of those regions’ components, placing the “Uralic” and “Eastern Mediterranean” components as much less relevant. If the K-means structure would have been analyzed to further depths, it’s possible that they had also detected other regional components (probaby a very diluted Italian-specific one I suspect must be there, hidden by the rest).
Anyhow, different studies (with different samples and different testing and statistical study means) produce somewhat different results and rather than any specific one, we should try to meta-read on all them according to their respective merits.
Terry: Autosomal PCs appear to be very susceptible to sampling and stuff like that. It’s quite interesting that CV in his book (not really a paper but a divulgation book, one of my first readings in genetics back there in the 90s, btw) finds a “Basque” component but, when you compare with other more recent studies, you realize that there are components (clusters) he did not detect (for instance a Central-North European one or an Iberian, non-Basque, one as well).
Also his attribution of some of the components appears misleading:
·PC1 and PC4: PC1 is attributed happily to Neolithic but in fact it hardly corresponds with it. PC4 instead does pretty well. PC1 might be Neolithic too but it appears to dominate areas where it’s not so strong actually, like Spain. PC4 in any case ahrdly relates to “Greek colonization” as Greeks never colonized the inland Balcans at all. That pattern appears much more closely related with Neolithic expansion also centered in Greece (Thessaly to be specific).
·PC2 and PC3: PC2 can ahrdly be considered Uralic once you look at the shadowed area in West Asia, where Uralic presence is unheard of. It rather seems to be some other kind of component, carried by Uralic peoples, sure, but also by other ethnicities, like Indo-Europeans maybe. The fact that it is much more important in terms of weight than PC3 also suggests that.
Anyhow PC1 and PC2 appear again in other studies (with some modifications) but when you look at deeper levels, they tend to become very thin in the West of Europe. They only appear that way because the Western diversity is greater and therefore the Western components appear more localized and weight less in the overall picture.
Check Bauchet et al, 2007 for a contrast. There you can see (K-means structure) how specific Central-North European and Iberian components appear and actually make up the biggest share of those regions’ components, placing the “Uralic” and “Eastern Mediterranean” components as much less relevant. If the K-means structure would have been analyzed to further depths, it’s possible that they had also detected other regional components (probaby a very diluted Italian-specific one I suspect must be there, hidden by the rest).
Anyhow, different studies (with different samples and different testing and statistical study means) produce somewhat different results and rather than any specific one, we should try to meta-read on all them according to their respective merits.
“you realize that there are components (clusters) he did not detect”. I think we can assume there have been dozens of movements into and around Europe over the years. Obviously these maps would pick up just five of them, not necessarily in order of magnitude but something approximating it.
“PC1 is attributed happily to Neolithic”. As you say, there are problems with this simple reading. It is much more likely to show the overall combined effect of all movements, as I pointed out in my original comment. I agree with your comments regarding PC4.
“PC2 can ahrdly be considered Uralic”. Totally agree. It’s something else, but what? Hardly likely to be Indo-European.
“Anyhow, different studies (with different samples and different testing and statistical study means) produce somewhat different results”. Basically the same trends though.
The next step would be to link these sorts of maps to similar work on crops and livestock: http://agro.biodiver.se/2007/10/linking-archaeology-and-genetics/
I’ve been looking at the DNA story of domesticates to try to nail down the origin of the Neolithic expansion. Turkey/Southern Turkey came up for all the earliest crops used in the West.
I can find nearly bugger all on far Eastern domesticates though. I’m guessing the work isn’t published in English